Riki Terui, Scott Berger, Larissa Sambel, Linda Song, and Gheorghe Chistol (Cell, 2024).
[Download PDF]
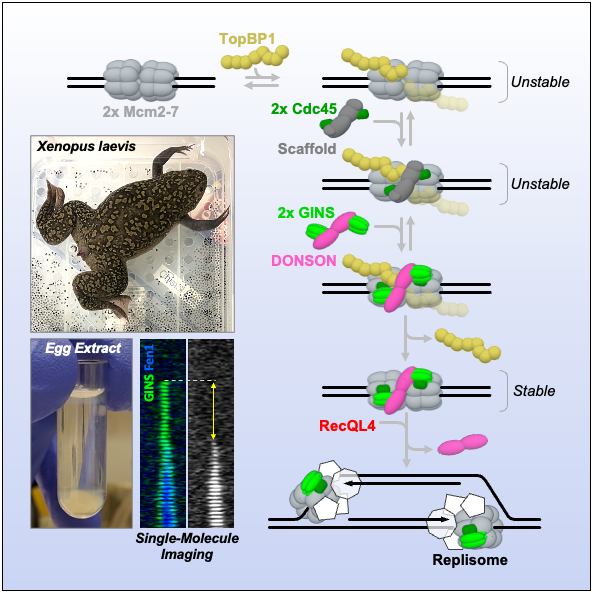
Metazoan genomes are copied bidirectionally from thousands of replication origins. Replication initiation entails the assembly and activation of two CMG helicases (Cdc45*Mcm2-7*GINS) at each origin. This requires several replication firing factors (including TopBP1, RecQL4, DONSON) whose exact roles are still under debate. How two helicases are correctly assembled and activated at each origin is a long-standing question. By visualizing the recruitment of GINS, Cdc45, TopBP1, RecQL4, and DONSON in real time, we uncovered that replication initiation is surprisingly dynamic. First, TopBP1 transiently binds to the origin and dissociates before the start of DNA synthesis. Second, two Cdc45 are recruited together, even though Cdc45 alone cannot dimerize. Next, two copies of DONSON and two GINS simultaneously arrive at the origin, completing the assembly of two CMG helicases. Finally, RecQL4 is recruited to the CMG*DONSON*DONSON*CMG complex and promotes DONSON dissociation and CMG activation via its ATPase activity.
Scott Berger and Gheorghe Chistol (Methods in Cell Biology, 2023).
[Download PDF]
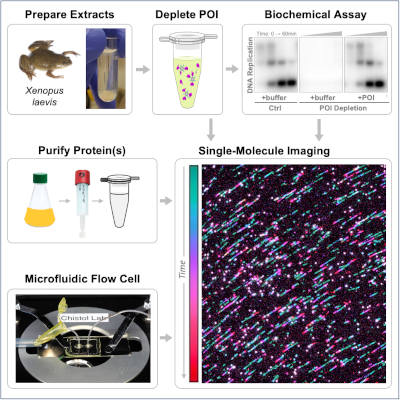
During cell division, the genome of each eukaryotic cell is copied by thousands of replisomes – large protein complexes consisting of several dozen proteins. Recent studies suggest that the eukaryotic replisome is much more dynamic than previously thought. To directly visualize replisome dynamics in a physiological context, we recently developed a single-molecule approach for imaging replication proteins in Xenopus egg extracts. These extracts contain all the soluble nuclear proteins and faithfully recapitulate DNA replication and repair in vitro, serving as a powerful platform for studying the mechanisms of genome maintenance. Here we present detailed protocols for conducting single-molecule experiments in nuclear egg extracts and preparing key reagents. This workflow can be easily adapted to visualize the dynamics and function of other proteins implicated in DNA replication and repair.
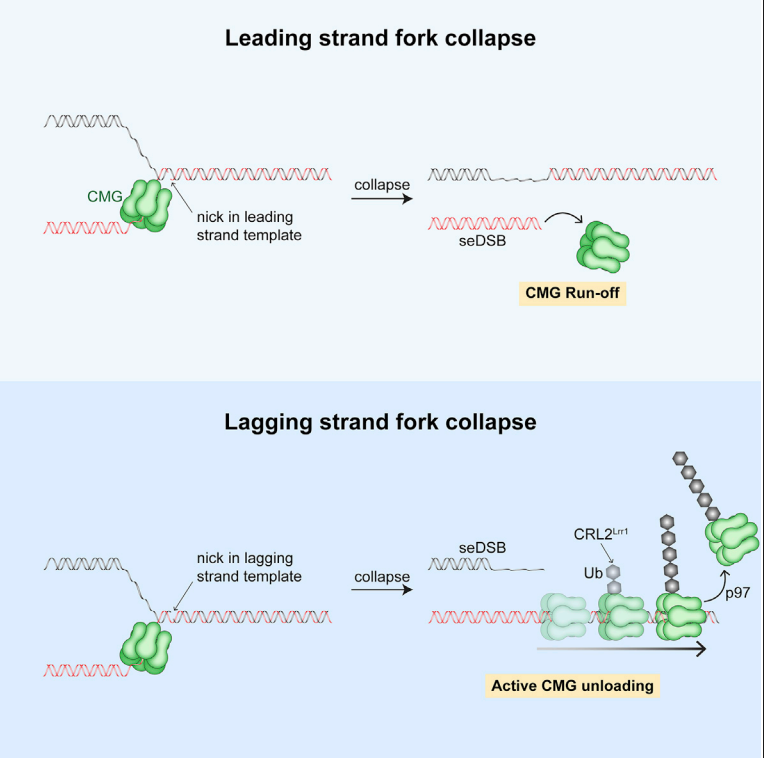
Vrtis K., Dewar J., Chistol G., Wu A.R., Graham T.G.W., Walter J.C. Molecular Cell, 81:1309-18 (2021). [PDF]
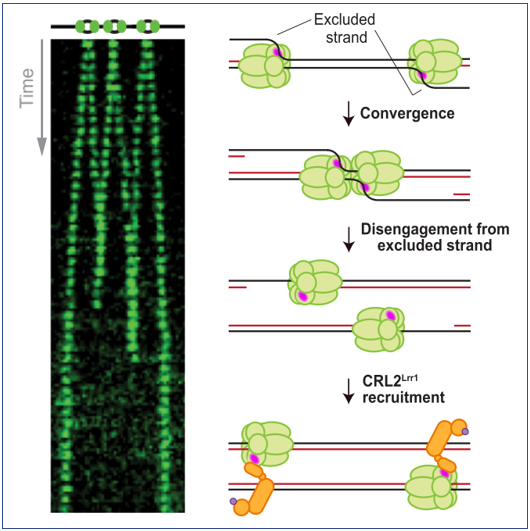
Low* E., Chistol* G [@]., Zaher M.S., Kochenova O.V., Walter J.C [@]. Genes & Development, 34:1534-45 (2020). [PDF]
* – these authors contributed equally.
[@] – corresponding authors
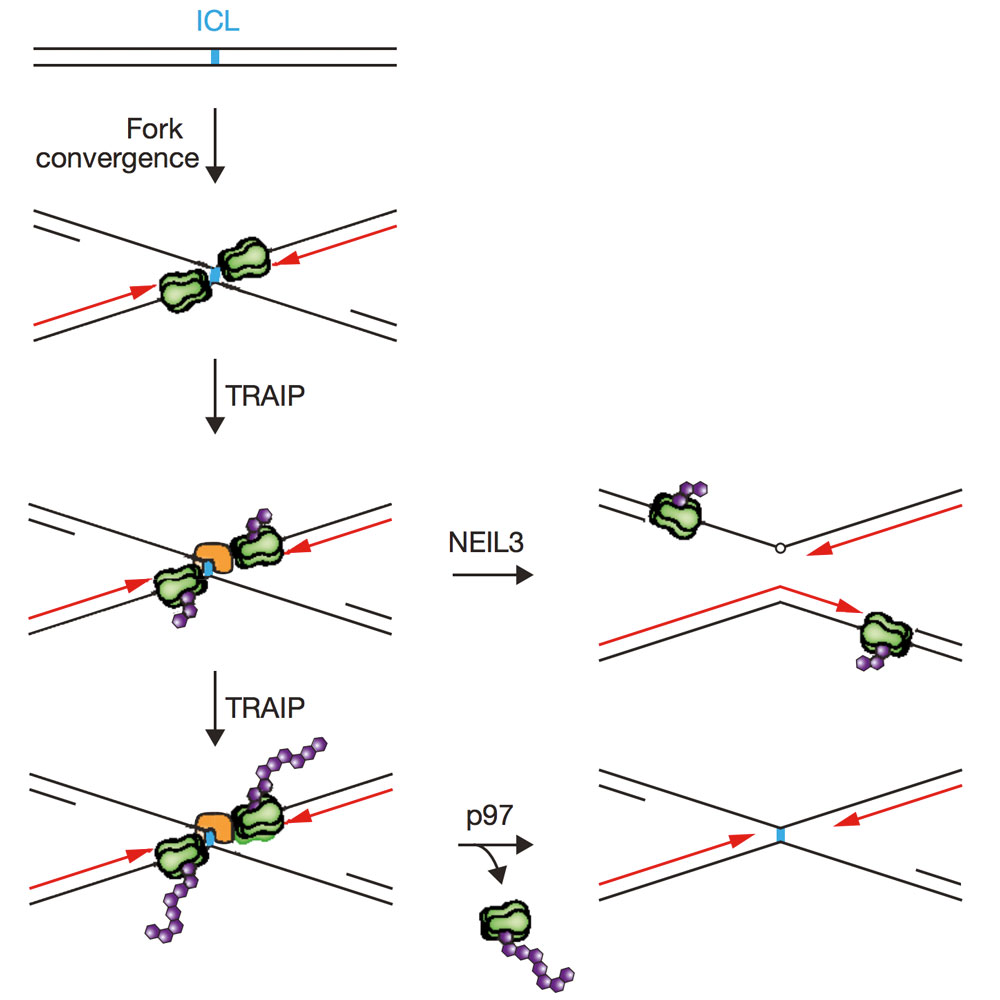
Wu R.A., Semlow D.R., Kamimae-Lanning A.N., Kochenova O.V., Chistol G. , Hodskinson M.R., Amunugama R., Sparks J.L., Wang M., Deng L., Mimoso C.A., Low E., Patel K.J., Walter J.C. Nature. 567(7747):267-72 (2019).
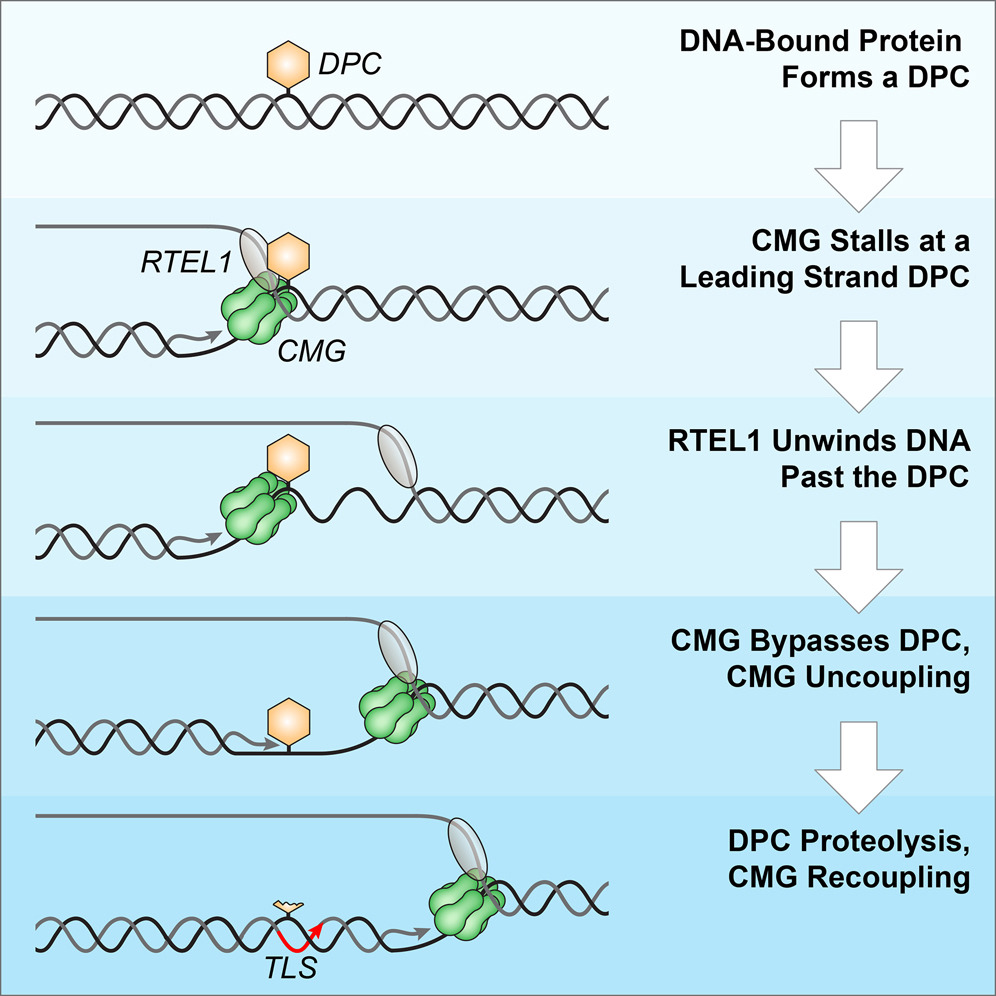
Sparks J.L.*, Chistol G.*, Gao A.O., Räschle M., Larsen N.B., Mann M., Duxin J.P., & Walter J.C. Cell, 176, 167-81.e21 (2019). [PDF]
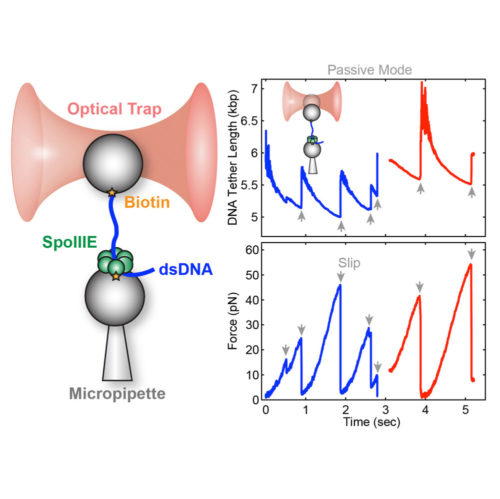
Liu N.*, Chistol G.*, Cui Y. & Bustamante C. eLife, 7 (2018). [PDF]
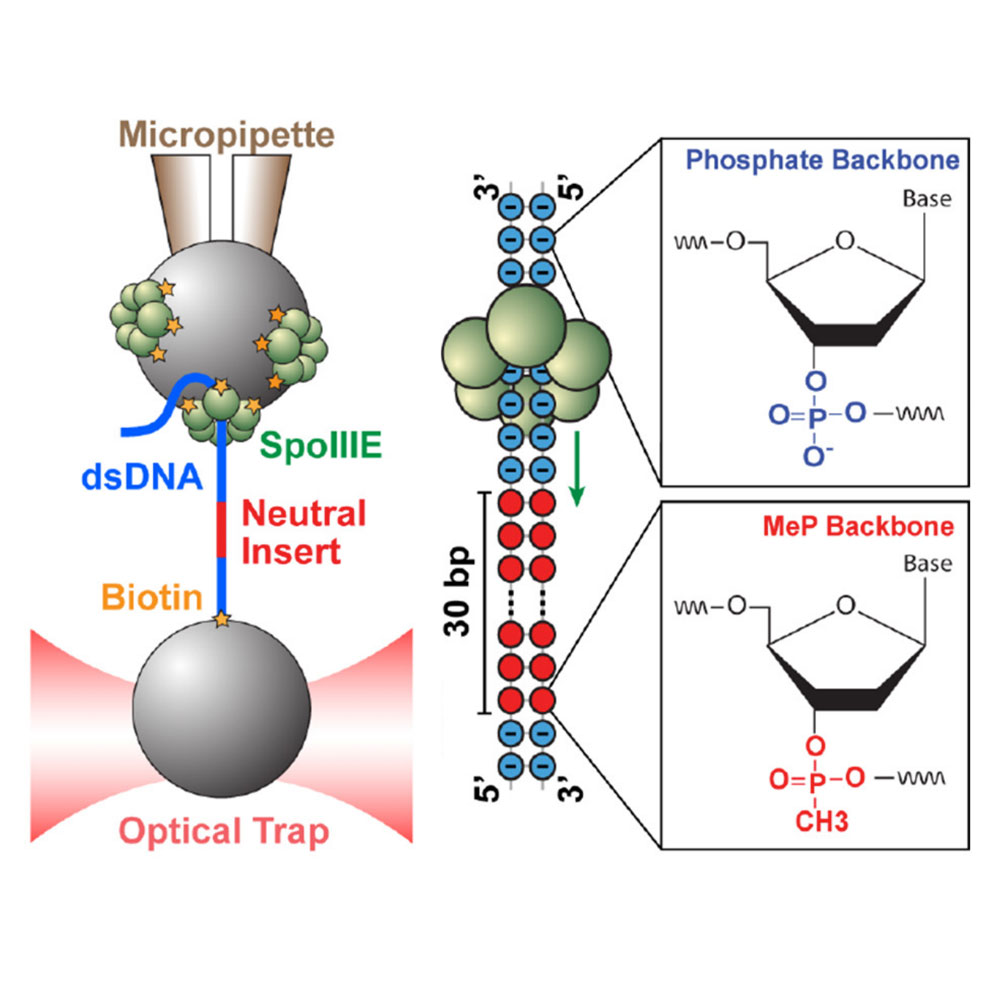
Liu N.*, Chistol G.* & Bustamante, C. eLife, 4 (2015). [PDF]
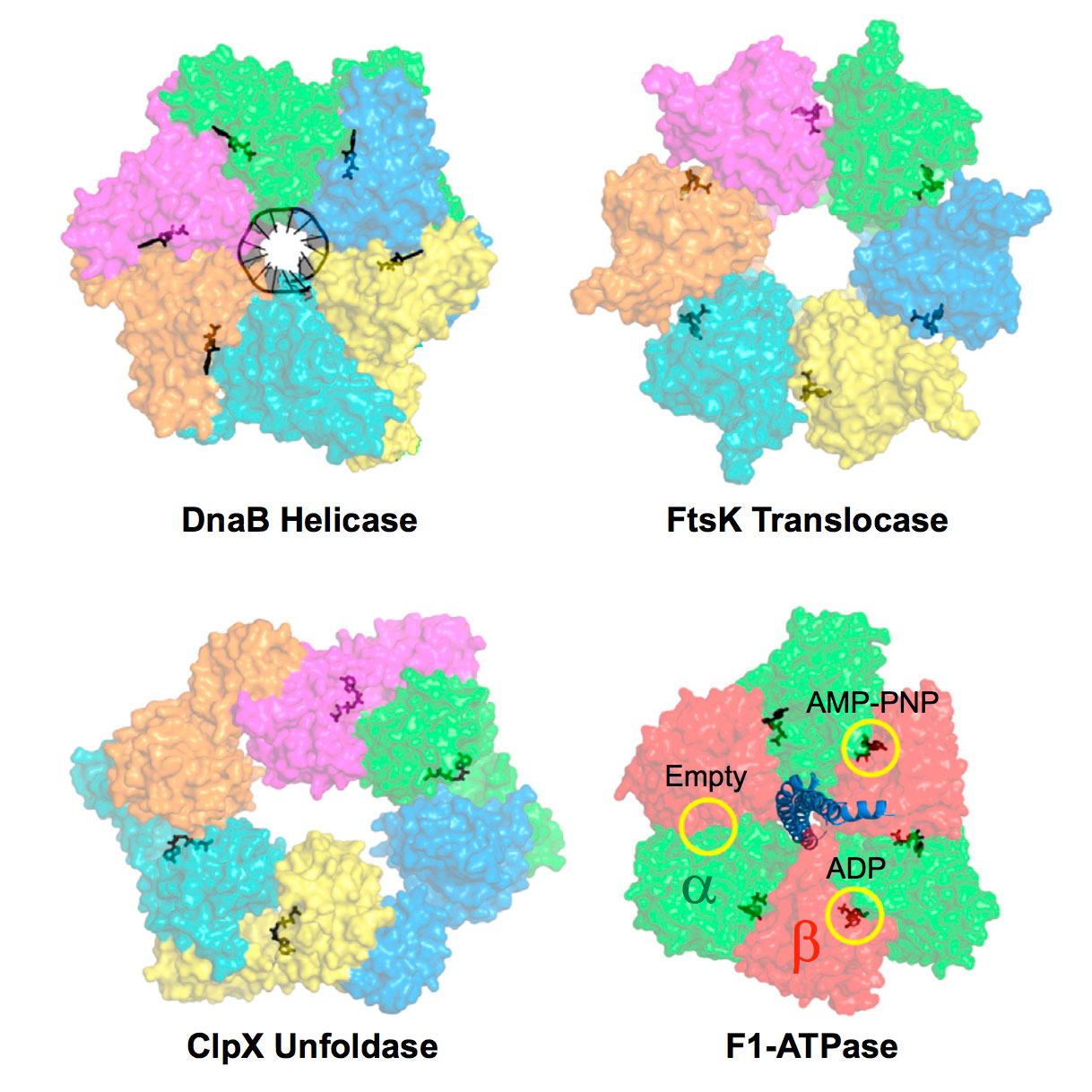
Liu S.*, Chistol G.* & Bustamante C. Biophysical Journal, 106, 1844–58 (2014). [PDF]
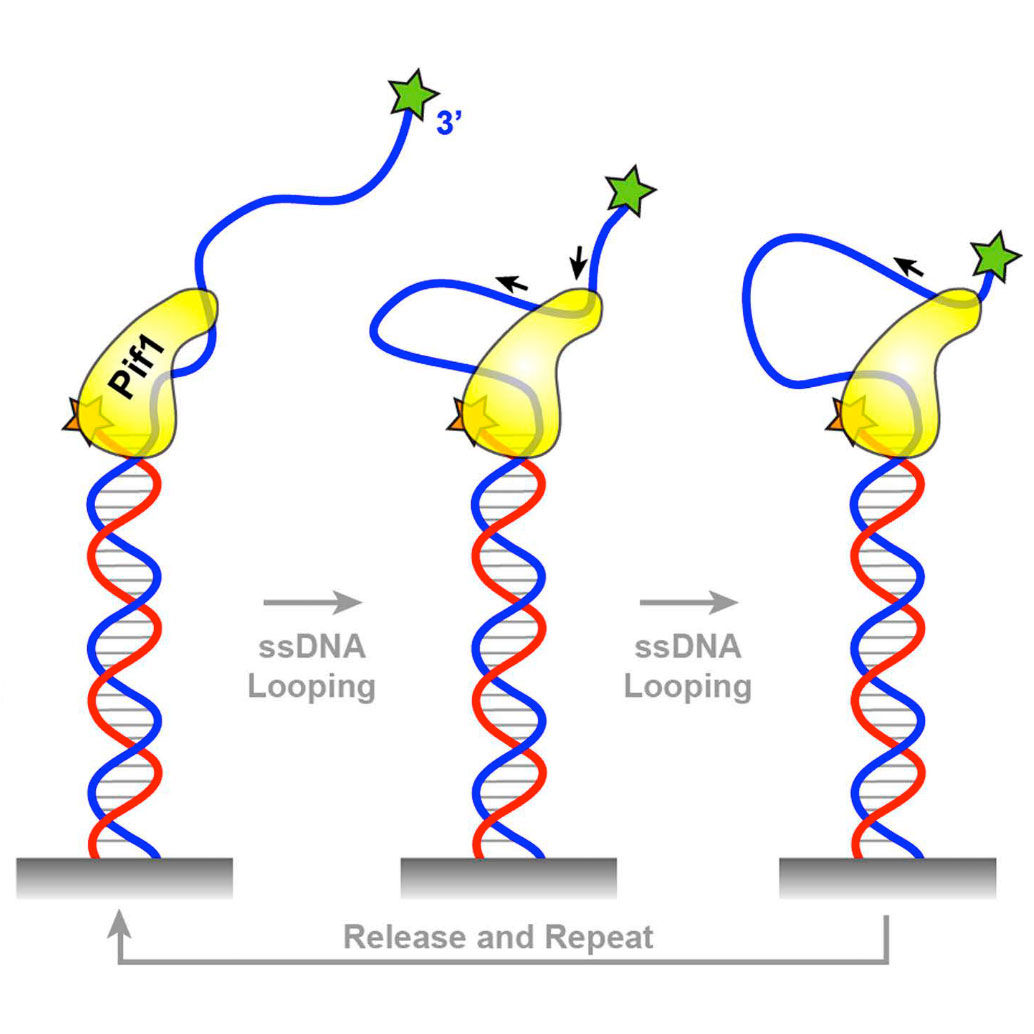
Chistol G. & Walter J.C. eLife 3, e02854 (2014). [PDF]
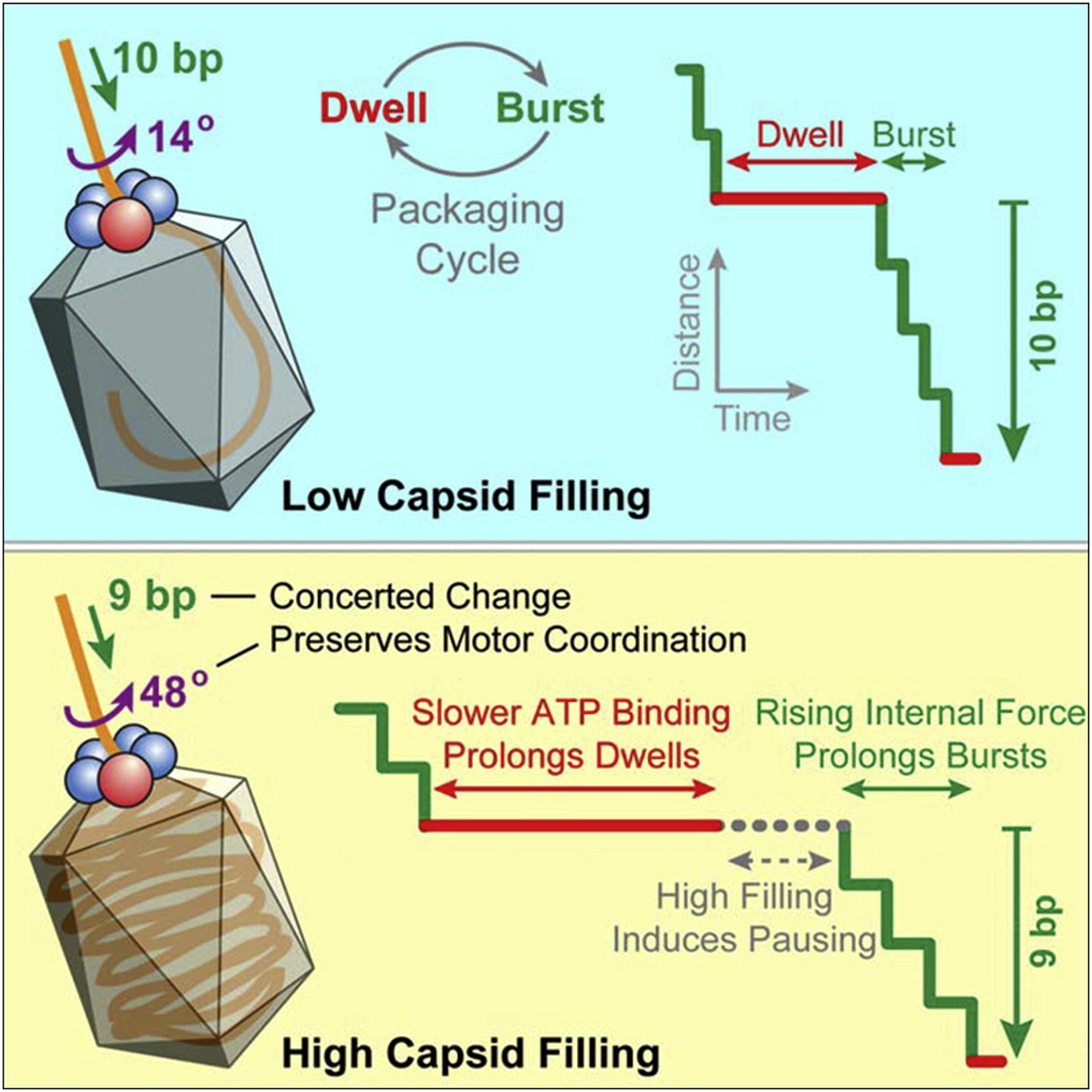
Liu S.*, Chistol G.*, Hetherington C.L.*, Tafoya S., Aathavan K., Schnitzbauer J., Grimes S., Jardine P.J., & Bustamante C. Cell 157, 702–13 (2014). [PDF]
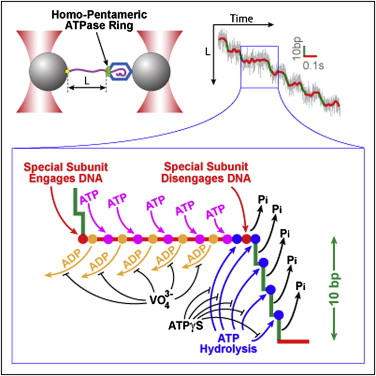
Chistol G.*, Liu S.*, Hetherington C.L., Moffitt J.R., Grimes S., Jardine P.J., & Bustamante C. Cell 151, 1017–28 (2012). [PDF]
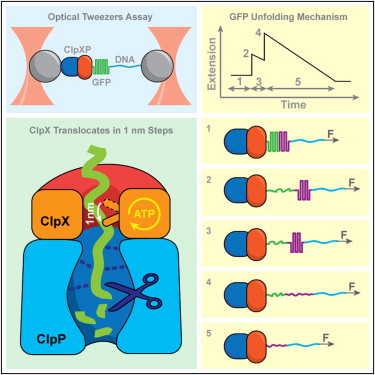
Maillard R.A., Chistol G., Sen M., Righini M., Tan J., Kaiser C.M., Hodges C., Martin A., & Bustamante C. Cell 145, 459–69 (2011). [PDF]
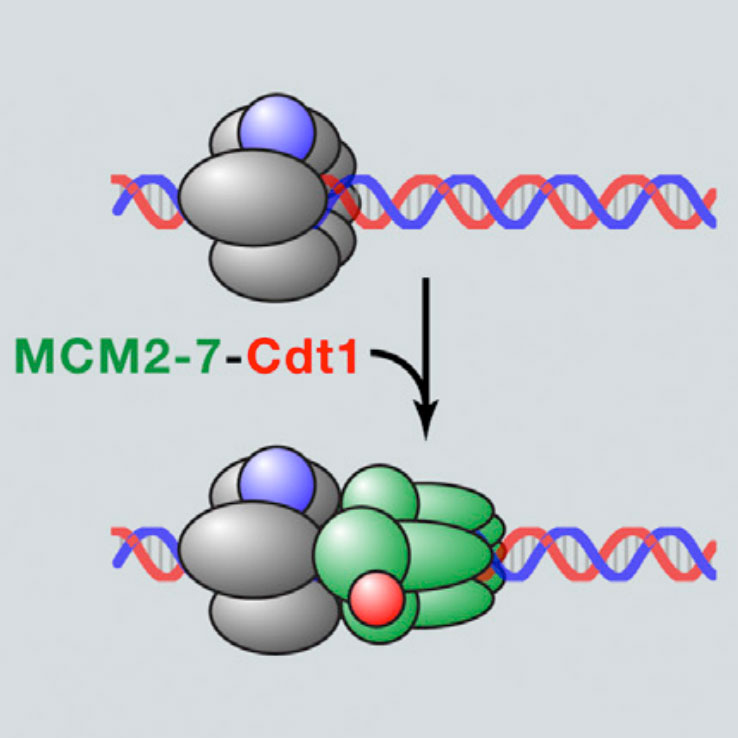
Chistol G. & Walter J.C. Cell 161, 429-30 (2015). [PDF]