To visualize the DNA replication machinery in real-time at the single-molecule level, we use several tools (click the links for more detail):
1. Linear DNA molecules are stretched and immobilized in a flow-cell.
2. The DNA is replicated in Xenopus Laevis nuclear egg extracts.
3. The replicative DNA helicase CMG is imaged using KEHRMIT.
4. The newly replicated DNA is imaged using PhADE.
5. Experiments are performed on a high-throughput TIRF microscope.
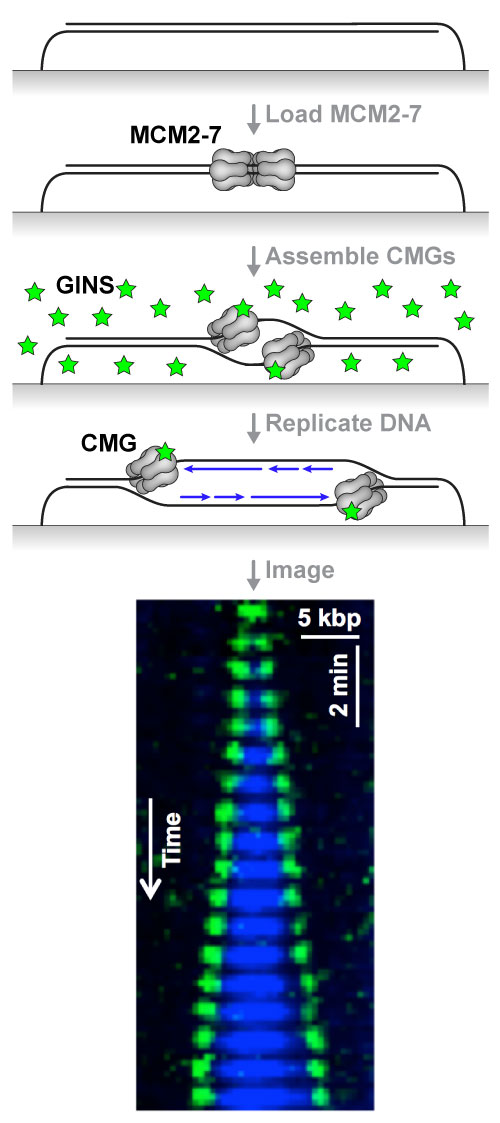
Attached is a diagram of the KEHRMIT+PhADE experiment: the inactive replicative helicase (MCM2-7 complex) is loaded onto immobilized DNA in HSS (an extract that mimics G1 phase); replication is initiated using NPE (an extract that mimics S phase) immuno-depleted of GINS (an essential helicase subunit) and supplemented with fluorescently-labeled recombinant GINS (green stars); excess free fluorescent protein is washed away and the DNA is replicated in nuclear egg extract. The kymogram shows two CMG helicases emerging from a replication origin (green spots, imaged by KEHRMIT), and the extent of the nascent DNA (blue tracts, imaged by PhADE). We are working to expand this single-molecule platform to enable direct imaging of many other essential genome maintenance factors whose function and regulation are poorly understood.
We aim to track the position of individual molecules (spatial information) in real-time (temporal information). In vivo eukaryotic genomes are highly compacted into chromatin – an unsuitable substrate for detecting the movement of genome maintenance proteins along DNA. To unlock the spatial information, we stretch linear DNA molecules (20-50 kb) using laminar flow, and immobilize them on a glass coverslip in a microfluidic flow-cell via biotin-streptavidin linkages. While conceptually similar to the DNA curtains method, this approach is technically much simpler. The attached movie shows Lambda DNA molecules (48.5 kb) immobilized at one or both ends and stained with a DNA-binding dye in the absence of any flow. The molecules tethered to the surface at both ends look like stretched ropes that flutter in the wind. Only the double-tethered DNA molecules are used for KEHRMIT or PhADE analysis.
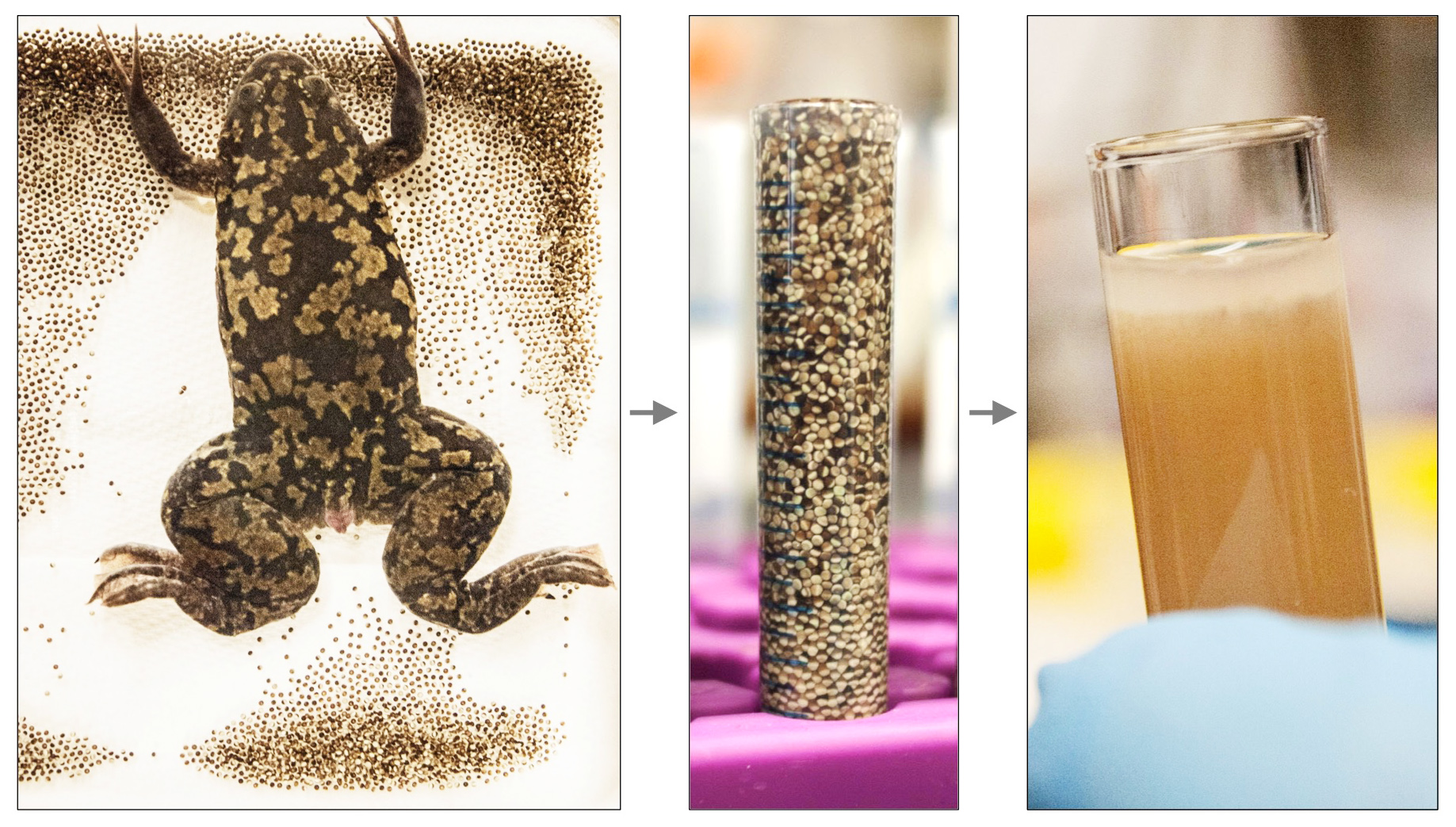
Xenopus egg extracts have been used for decades to study various aspects of the cell cycle. Several extract “recipes” exist: crude extract, Low-Speed Supernatant (LSS), High-Speed Supernatant (HSS), etc. As a postdoc in the Newport lab at UCSD, Johannes Walter developed the nucleoplasmic extract (NPE) – a cell-free, membrane-free extract highly-enriched in nuclear factors. NPE contains all soluble frog proteins and efficiently recapitulates DNA replication, all major DNA repair pathways, and many regulatory aspects of genome maintenance. We use HSS and NPE to mimic the G1 and S phases of the cell cycle respectively, and replicate/repair any DNA substrate of interest, including artificially-constructed molecules that bear pre-defined sequences or specific forms of DNA damage.
As a postdoc in Johannes Walter’s lab at Harvard Medical School, Gheorghe Chistol developed KEHRMIT (Kinetics of the Eukaryotic Helicase by Real-time Molecular Imaging and Tracking) – a novel single-molecule technique to directly visualize the activity of the replicative DNA helicase. In KEHRMIT, stretched DNA is replicated in extracts supplemented with a fluorescent subunit (GINS) that is incorporated into each helicase – a key replisome component. KEHRMIT was first used to dissect how the replisome copes with DNA-protein crosslinks (DPCs) – a very toxic form of DNA damage in which a protein is covalently attached to a DNA strand. Attached is a sped-up movie from a 1-hr KEHRMIT experiment where each helicase appears as a diffraction-limited spot. Two helicases emanate from each replication origin, and each helicase molecule functions within a replisome – a large multi-subunit molecular complex tasked with replicating DNA.
To visualize nascent DNA, we employ PhADE – a single-molecule approach developed by Anna Loveland in the Walter and van Oijen labs at Harvard Medical School. The extract is supplemented with recombinant Fen1 (an enzyme that binds Okazaki fragments and cleaves 5′-flaps) fused to mKikGR – a green fluorescent protein that upon exposure to UV light becomes an orange/red fluorescent protein. PhADE entails (1) illuminating the sample with UV in TIRF mode so only Fen1-mKikGR molecules within ~100nm of the flow cell surface are photo-switched; (2) allowing any free photo-switched Fen1-mKikGR to diffuse away; and (3) TIRF-imaging any photo-switched Fen1-mKikGR that remains near the surface because it is bound to nascent DNA. The beauty of this approach is that it enables single-molecule detection with good signal-to-noise even though Fen1-mKikGR is present at huge concentrations (1-5uM), 100-1000x higher than those compatible with conventional single-molecule imaging.
We currently use a commercial Nikon Eclipse Ti2 equipped with a 4-laser light source (405, 488, 561, 640 nm), a high numerical aperture 100x TIRF objective, a motorized TIRF illuminator for reproducible TIRF angle control, an autofocus system, a fast motorized stage, two Andor iXon Ultra 897 EMCCD cameras with a Cairn TwinCam image splitter for simultaneous full-frame two-color imaging. The combination of objective+camera is capable of imaging an 82×82 micron field of view at any given time. As illustrated in the attached movie, in one experiment we periodically sample several (50-100) fields of view over the course of hours, dramatically increasing the throughput of our single-molecule KEHRMIT+PhADE assay.