The DNA replication fork suppresses CMG unloading from chromatin before termination
September 21, 2020
 Low* E., Chistol* G [@]., Zaher M.S., Kochenova O.V., Walter J.C [@]. Genes & Development, 34:1-12 (2020). [PDF]
Low* E., Chistol* G [@]., Zaher M.S., Kochenova O.V., Walter J.C [@]. Genes & Development, 34:1-12 (2020). [PDF]
* – these authors contributed equally.
[@] – corresponding authors
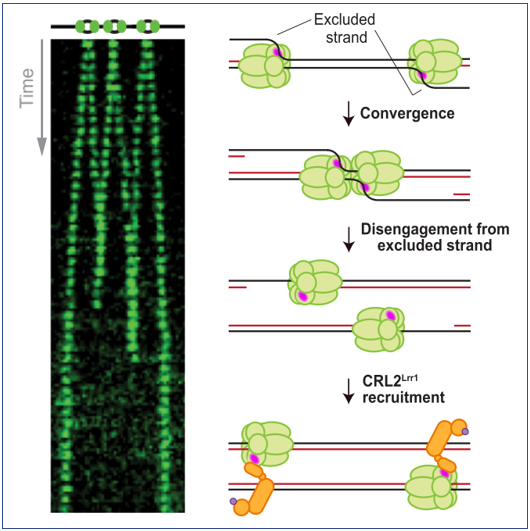
When converging replication forks meet during replication termination, the CMG (Cdc45–MCM2–7–GINS) helicase is polyubiquitylated by CRL2Lrr1 and unloaded from chromatin by the p97 ATPase. Here, we investigate the signal that triggers CMG unloading in Xenopus egg extracts using single-molecule and ensemble approaches. We show that converging CMGs pass each other and keep translocating at the same speed as before convergence, whereafter they are rapidly and independently unloaded. When CMG unloading is blocked, diverging CMGs do not support DNA synthesis, indicating that after bypass CMGs encounter the nascent lagging strands of the converging fork and then translocate along double-stranded DNA (dsDNA). However, translocation on dsDNA is not required for CMG’s removal from chromatin because in the absence of nascent strand synthesis, converging CMGs are still unloaded. Moreover, recombinant CMG added to nuclear extract undergoes ubiquitylation and disassembly in the absence of any DNA, and DNA digestion triggers CMG ubiquitylation at stalled replication forks. Our findings suggest that DNA suppresses CMG ubiquitylation during elongation and that this suppression is relieved when CMGs converge, leading to CMG unloading.
